Is The Nucleolus Found In Plants Or Animals
| Prison cell biological science | |
|---|---|
| Animal cell diagram | |
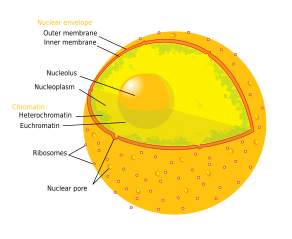
 Components of a typical beast cell:
|
The nucleolus (, plural: nucleoli ) is the largest structure in the nucleus of eukaryotic cells.[1] It is best known as the site of ribosome biogenesis. Nucleoli also participate in the formation of bespeak recognition particles and play a office in the cell'southward response to stress.[two] Nucleoli are fabricated of proteins, DNA and RNA and grade around specific chromosomal regions called nucleolar organizing regions. Malfunction of nucleoli tin can be the crusade of several human weather chosen "nucleolopathies"[iii] and the nucleolus is being investigated as a target for cancer chemotherapy.[4] [five]
History [edit]
The nucleolus was identified by brilliant-field microscopy during the 1830s.[half-dozen] Little was known near the function of the nucleolus until 1964, when a study[7] of nucleoli by John Gurdon and Donald Brown in the African clawed frog Xenopus laevis generated increasing interest in the function and detailed construction of the nucleolus. They found that 25% of the frog eggs had no nucleolus and that such eggs were not capable of life. Half of the eggs had 1 nucleolus and 25% had two. They concluded that the nucleolus had a role necessary for life. In 1966 Max L. Birnstiel and collaborators showed via nucleic acid hybridization experiments that Dna within nucleoli code for ribosomal RNA.[eight] [9]
Construction [edit]
Three major components of the nucleolus are recognized: the fibrillar centre (FC), the dense fibrillar component (DFC), and the granular component (GC).[1] Transcription of the rDNA occurs in the FC.[ten] The DFC contains the protein fibrillarin,[10] which is important in rRNA processing. The GC contains the poly peptide nucleophosmin,[10] (B23 in the external prototype) which is besides involved in ribosome biogenesis.
However, information technology has been proposed that this particular organization is only observed in higher eukaryotes and that it evolved from a bipartite organization with the transition from anamniotes to amniotes. Reflecting the substantial increase in the Deoxyribonucleic acid intergenic region, an original fibrillar component would have separated into the FC and the DFC.[xi]
Nucleus from a prison cell line. Fibrillarin in red. Transcription regulatory protein CTCFL in green. Nuclear DNA in bluish.
Another construction identified within many nucleoli (particularly in plants) is a clear surface area in the center of the structure referred to every bit a nucleolar vacuole.[12] Nucleoli of diverse plant species have been shown to have very loftier concentrations of iron[13] in contrast to human and animal jail cell nucleoli.
The nucleolus ultrastructure can be seen through an electron microscope, while the system and dynamics can exist studied through fluorescent protein tagging and fluorescent recovery later photobleaching (FRAP). Antibodies against the PAF49 protein can also exist used as a marker for the nucleolus in immunofluorescence experiments.[14]
Although normally only one or ii nucleoli can be seen, a diploid human jail cell has ten nucleolus organizer regions (NORs) and could have more nucleoli. Most often multiple NORs participate in each nucleolus.[15]
Function and ribosome assembly [edit]

Electron micrograph of part of a HeLa prison cell. The paradigm is a screen capture from this moving picture, which shows a Z-stack of the prison cell.
In ribosome biogenesis, two of the three eukaryotic RNA polymerases (pol I and Three) are required, and these part in a coordinated manner. In an initial stage, the rRNA genes are transcribed every bit a unmarried unit inside the nucleolus past RNA polymerase I. In order for this transcription to occur, several pol I-associated factors and DNA-specific trans-acting factors are required. In yeast, the near of import are: UAF (upstream activating factor), TBP (TATA-box binding protein), and cadre binding cistron (CBF)) which demark promoter elements and form the preinitiation complex (PIC), which is in turn recognized by RNA pol. In humans, a similar PIC is assembled with SL1, the promoter selectivity factor (composed of TBP and TBP-associated factors, or TAFs), transcription initiation factors, and UBF (upstream binding factor). RNA polymerase I transcribes nearly rRNA transcripts (28S, 18S, and 5.8S) merely the 5S rRNA subunit (component of the 60S ribosomal subunit) is transcribed past RNA polymerase III.[16]
Transcription of rRNA yields a long precursor molecule (45S pre-rRNA) which notwithstanding contains the ITS and ETS. Farther processing is needed to generate the 18S RNA, 5.8S and 28S RNA molecules. In eukaryotes, the RNA-modifying enzymes are brought to their respective recognition sites by interaction with guide RNAs, which bind these specific sequences. These guide RNAs belong to the course of minor nucleolar RNAs (snoRNAs) which are complexed with proteins and exist as small-nucleolar-ribonucleoproteins (snoRNPs). Once the rRNA subunits are processed, they are set to exist assembled into larger ribosomal subunits. However, an additional rRNA molecule, the 5S rRNA, is besides necessary. In yeast, the 5S rDNA sequence is localized in the intergenic spacer and is transcribed in the nucleolus past RNA pol.
In college eukaryotes and plants, the situation is more than complex, for the 5S DNA sequence lies outside the NOR and is transcribed by RNA pol III in the nucleoplasm, after which it finds its way into the nucleolus to participate in the ribosome assembly. This assembly not only involves the rRNA, but ribosomal proteins besides. The genes encoding these r-proteins are transcribed by pol II in the nucleoplasm by a "conventional" pathway of poly peptide synthesis (transcription, pre-mRNA processing, nuclear export of mature mRNA and translation on cytoplasmic ribosomes). The mature r-proteins are and then imported into the nucleus and finally the nucleolus. Association and maturation of rRNA and r-proteins upshot in the formation of the 40S (small) and 60S (large) subunits of the consummate ribosome. These are exported through the nuclear pore complexes to the cytoplasm, where they remain free or become associated with the endoplasmic reticulum, forming rough endoplasmic reticulum (RER).[17] [18]
In human endometrial cells, a network of nucleolar channels is sometimes formed. The origin and function of this network has not yet been conspicuously identified.[xix]
Sequestration of proteins [edit]
In addition to its role in ribosomal biogenesis, the nucleolus is known to capture and immobilize proteins, a process known equally nucleolar detention. Proteins that are detained in the nucleolus are unable to diffuse and to interact with their binding partners. Targets of this mail service-translational regulatory machinery include VHL, PML, MDM2, POLD1, RelA, HAND1 and hTERT, among many others. It is now known that long noncoding RNAs originating from intergenic regions of the nucleolus are responsible for this miracle.[20]
See also [edit]
- Differential interference contrast microscopy
References [edit]
- ^ a b O'Sullivan JM, Pai DA, Cridge AG, Engelke DR, Ganley AR (June 2013). "The nucleolus: a raft adrift in the nuclear sea or the keystone in nuclear construction?". Biomolecular Concepts. 4 (3): 277–86. doi:10.1515/bmc-2012-0043. PMC5100006. PMID 25436580.
- ^ Olson MO, Dundr Yard (16 Feb 2015). "Nucleolus: Structure and Function". Encyclopedia of Life Sciences (eLS). doi:10.1002/9780470015902.a0005975.pub3. ISBN978-0-470-01617-6.
- ^ Hetman M (June 2014). "Part of the nucleolus in human diseases. Preface". Biochimica et Biophysica Acta. 1842 (vi): 757. doi:10.1016/j.bbadis.2014.03.004. PMID 24631655.
- ^ Quin JE, Devlin JR, Cameron D, Hannan KM, Pearson RB, Hannan RD (June 2014). "Targeting the nucleolus for cancer intervention". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Illness. 1842 (vi): 802–16. doi:10.1016/j.bbadis.2013.12.009. PMID 24389329.
- ^ Woods SJ, Hannan KM, Pearson RB, Hannan RD (July 2015). "The nucleolus as a fundamental regulator of the p53 response and a new target for cancer therapy". Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 1849 (7): 821–9. doi:10.1016/j.bbagrm.2014.x.007. PMID 25464032.
- ^ Pederson T (March 2011). "The nucleolus". Cold Jump Harbor Perspectives in Biology. 3 (three): a000638. doi:x.1101/cshperspect.a000638. PMC3039934. PMID 21106648.
- ^ Brown DD, Gurdon JB (Jan 1964). "Absence of ribosomal rna synthesis in the anucleolate mutant of xenopus laevis". Proceedings of the National Academy of Sciences of the Us of America. 51 (1): 139–46. Bibcode:1964PNAS...51..139B. doi:10.1073/pnas.51.1.139. PMC300879. PMID 14106673.
- ^ Birnstiel ML, Wallace H, Sirlin JL, Fischberg M (December 1966). "Localization of the ribosomal DNA complements in the nucleolar organizer region of Xenopus laevis". National Cancer Institute Monograph. 23: 431–47. PMID 5963987.
- ^ Wallace H, Birnstiel ML (Feb 1966). "Ribosomal cistrons and the nucleolar organizer". Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Poly peptide Synthesis. 114 (2): 296–310. doi:10.1016/0005-2787(66)90311-10. PMID 5943882.
- ^ a b c Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D (Jan 2008). "Nucleolus: the fascinating nuclear trunk". Histochemistry and Cell Biology. 129 (1): 13–31. doi:10.1007/s00418-007-0359-half dozen. PMC2137947. PMID 18046571.
- ^ Thiry Chiliad, Lafontaine DL (April 2005). "Nativity of a nucleolus: the evolution of nucleolar compartments". Trends in Cell Biology. fifteen (4): 194–9. doi:x.1016/j.tcb.2005.02.007. PMID 15817375. as PDF Archived 17 December 2008 at the Wayback Machine
- ^ Beven AF, Lee R, Razaz M, Leader DJ, Brownish JW, Shaw PJ (June 1996). "The organisation of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs". Journal of Cell Science. 109 ( Pt 6) (6): 1241–51. doi:10.1242/jcs.109.half dozen.1241. PMID 8799814.
- ^ Roschzttardtz H, Grillet Fifty, Isaure MP, Conéjéro G, Ortega R, Curie C, Mari S (Baronial 2011). "Plant jail cell nucleolus as a hot spot for fe". The Journal of Biological Chemical science. 286 (32): 27863–6. doi:ten.1074/jbc.C111.269720. PMC3151030. PMID 21719700.
- ^ PAF49 antibody | GeneTex Inc. Genetex.com. Retrieved 2019-07-eighteen.
- ^ von Knebel Doeberitz G, Wentzensen N (2008). "The Jail cell: Basic Construction and Function". Comprehensive Cytopathology (third ed.).
- ^ Champe PC, Harvey RA, Ferrier DR (2005). Lippincott's Illustrated Reviews: Biochemistry. Lippincott Williams & Wilkins. ISBN978-0-7817-2265-0.
- ^ Alberts B, Johnson A, Lewis J, Raff Chiliad, Roberts Yard, Walter P (2002). Molecular Biology of the Cell (4th ed.). New York: Garland Science. pp. 331–three. ISBN978-0-8153-3218-3.
- ^ Cooper GM, Hausman RE (2007). The Jail cell: A Molecular Approach (4th ed.). Sinauer Associates. pp. 371–9. ISBN978-0-87893-220-7.
- ^ Wang T, Schneider J (i July 1992). "Origin and fate of the nucleolar channel system of normal man endometrium". Jail cell Research. two (2): 97–102. doi:x.1038/cr.1992.10.
- ^ Audas TE, Jacob Medico, Lee S (Jan 2012). "Immobilization of proteins in the nucleolus past ribosomal intergenic spacer noncoding RNA". Molecular Prison cell. 45 (2): 147–57. doi:10.1016/j.molcel.2011.12.012. PMID 22284675.
Further reading [edit]
- Cooper GM (2000). "The Nucleolus". The Cell: A Molecular Approach (2nd ed.). Sunderland MA: Sinauer Associates. ISBN978-0-87893-106-4.
- Tiku 5, Antebi A (August 2018). "Nucleolar Function in Lifespan Regulation". Trends in Jail cell Biological science. 28 (8): 662–672. doi:10.1016/j.tcb.2018.03.007. PMID 29779866. S2CID 29167518.
- JoAnna Klein (20 May 2018). "The Thing Inside Your Cells That Might Make up one's mind How Long You Live". The New York Times.
External links [edit]
| | Look upwards nucleolus in Wiktionary, the complimentary dictionary. |
- Nucleolus nether electron microscope II at uni-mainz.de
- Nuclear Poly peptide Database – search nether compartment
- Cell+Nucleolus at the US National Library of Medicine Medical Subject area Headings (MeSH)
- Histology image: 20104loa – Histology Learning Organization at Boston University
Source: https://en.wikipedia.org/wiki/Nucleolus
Posted by: browntheird.blogspot.com

0 Response to "Is The Nucleolus Found In Plants Or Animals"
Post a Comment